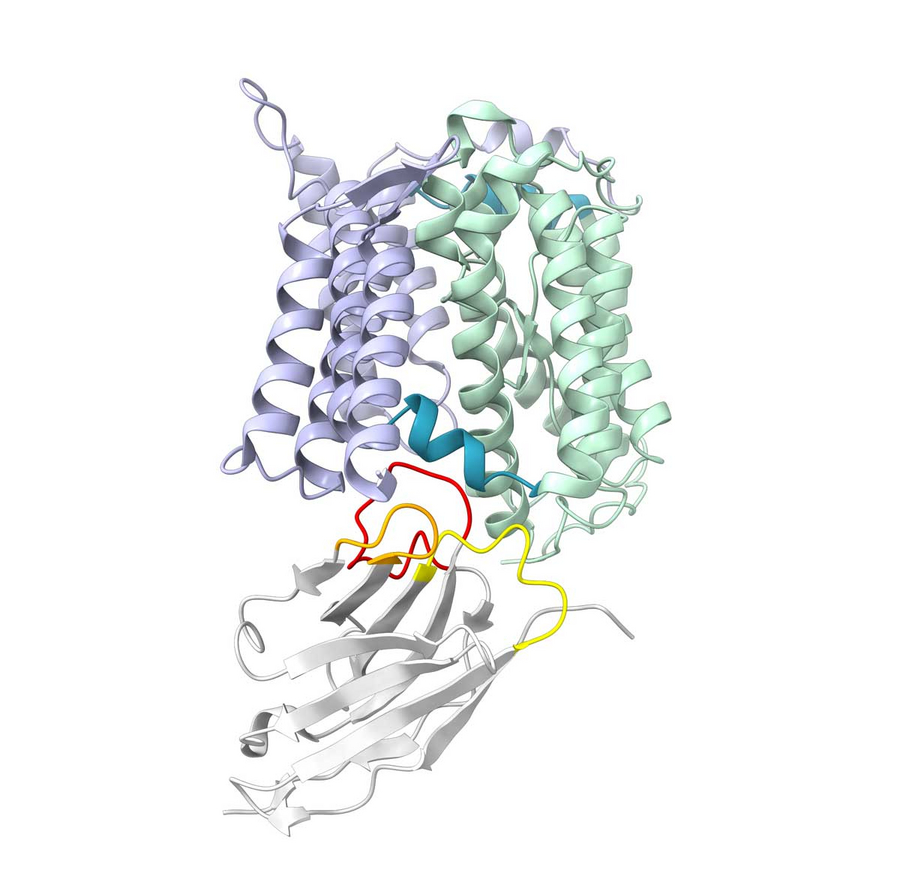
Protein structure displaying the hinge regions (blue) that connect the static (purple) and mobile (green) parts of an elevator transporter. A nanobody (grey) locks the protein in the ‘down’ position. Copyright: Kuhn et al. / Nat. Commun. (2024) / MPI-CBG
Biological membranes surround all cells of all living organisms. Many molecules cannot freely pass through these membranes, even though cells need them. To survive, cells therefore make use of complex membrane transport machinery to take up, for example, nutrients from their environment. These machines are membrane transport proteins. The largest family of membrane transport proteins are called solute carriers (SLCs). Several members of this family use a mechanism of transport that resembles an elevator: a mobile part of the protein slides along a static part of the protein from the outside of the membrane to the inside of the cell, transporting various essential molecules. But how this movement was coordinated had remained unclear until now. A team of researchers, led by Eric Geertsma at the Max Planck Institute of Molecular Cell Biology (MPI-CBG) in Dresden and previously at the Goethe University Frankfurt, together with collaborators from the Max Planck Institute for Brain Research, the Max Planck Institute of Biophysics, and the Institute of Medical Microbiology at the University of Zurich, now describe an overlooked structural element in these elevators that functions as a hinge between the mobile and static part. This hinge regulates the elevator’s movements and thereby its function. This uncovers another aspect of the underlying mechanism of the protein family of elevator transporters.
Elevator dynamics
The team’s attention was drawn to this part of the protein because it had always been overlooked. “We noticed that no one ever studied the relevance of this part of the protein, though nearly all elevator proteins carry it. That made it interesting for us,” explains Benedikt Kuhn, the first author of the study. They performed their studies on the SLC23 family, whose human members transport Vitamin C. To understand how the hinge affects the transporter’s dynamics, the researchers created single amino acid variations, so-called mutations. They then compared the behavior of the altered proteins with the original protein, using probes that indicate whether the elevator is up or down. They found that single mutations in the hinge region could change the resting position of the elevator from ‘down’, accessing the inside of the cell, to ‘up’, accessing the outside of the cell, or to a position in between. Importantly, these mutations dramatically affected the transport function as well.
Wedging the door
To determine whether an elevator that is up can still go down, the researchers used a nanobody, which is a small piece of camelid antibodies, that specifically binds to the ‘elevator down’ state and locks it in place, much like a wedge in a door. Indeed, the elevators that were mostly up due to the hinge mutation could be trapped by the nanobody in the down state. This is important, as while the previous experiments only showed the position of the elevator, this experiment confirmed it could still move down. The elevator with the other mutation, that remains in the middle, was not trapped by the nanobody and thus does not move fully down. Different mutations in the hinge can therefore completely change the elevator’s movements, underscoring the importance of the hinge.
Eric Geertsma, who supervised the study, concludes: “The discovery not only provides important fundamental knowledge on how transport dynamics of this family of membrane proteins are organized. It provides new leads that could aid in the re-activation of defective proteins in disease.”
Kuhn, B.T., Zöller, J., Zimmermann, I. et al. Interdomain-linkers control conformational transitions in the SLC23 elevator transporter UraA. Nat Commun 15, 7518 (2024). https://doi.org/10.1038/s41467-024-51814-8